Difference between revisions of "Statistical Algorithms Importer: Pre-Installed Project FAQ"
(→How to use the input and output parameters) |
|||
| (26 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
|} | |} | ||
| − | F.A.Q. of Statistical Algorithms Importer (SAI), here are common mistakes we have found in Pre-Installed Project. | + | F.A.Q. of [[Statistical_Algorithms_Importer|Statistical Algorithms Importer (SAI)]], here are common mistakes we have found in Pre-Installed Project. |
| Line 12: | Line 12: | ||
To solve this problem you can use the "convert to unix" functionality provided by notepad++ for example, or look for this feature in your editor. | To solve this problem you can use the "convert to unix" functionality provided by notepad++ for example, or look for this feature in your editor. | ||
| − | == How | + | == How to use the input and output parameters == |
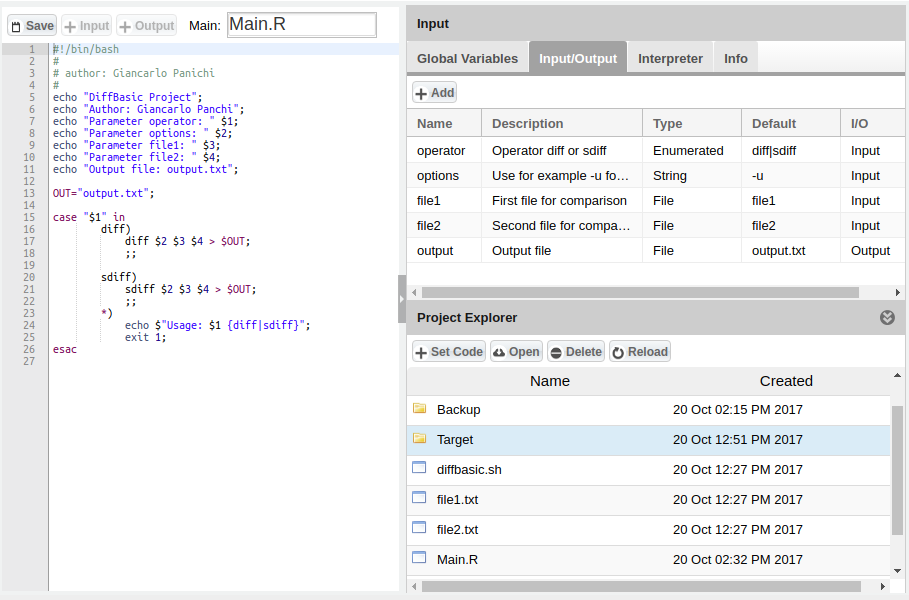
| − | : | + | :For example we consider DiffBasic algorithm: |
| − | [[Image:StatisticalAlgorithmsImporter_DiffBasic0.png|thumb|center| | + | [[Image:StatisticalAlgorithmsImporter_DiffBasic0.png|thumb|center|750px|DiffBasic, SAI]] |
| + | |||
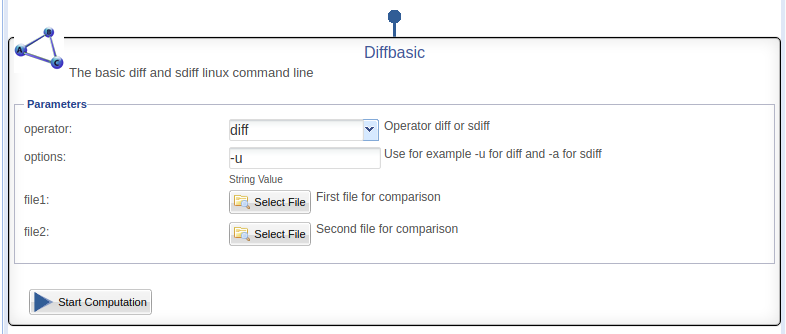
| + | :DataMiner show the DiffBasic algorithm in this way: | ||
| + | [[Image:StatisticalAlgorithmsImporter_DiffBasic1.png|thumb|center|750px|DiffBasic in DataMiner interface, SAI]] | ||
:Bash script code in sample: | :Bash script code in sample: | ||
| + | <source lang='bash'> | ||
| + | #!/bin/bash | ||
| + | # | ||
| + | # author: Giancarlo Panichi | ||
| + | # | ||
| + | echo "DiffBasic Project" | ||
| + | echo "Author: Giancarlo Panchi" | ||
| + | echo "Parameter operator: " $1 | ||
| + | echo "Parameter options: " $2 | ||
| + | echo "Parameter file1: " $3 | ||
| + | echo "Parameter file2: " $4 | ||
| + | echo "Output file: output.txt" | ||
| + | OUT="output.txt" | ||
| + | |||
| + | case "$1" in | ||
| + | diff) | ||
| + | diff $2 $3 $4 > $OUT | ||
| + | ;; | ||
| + | |||
| + | sdiff) | ||
| + | sdiff $2 $3 $4 > $OUT | ||
| + | ;; | ||
| + | *) | ||
| + | echo $"Usage: $1 {diff|sdiff}" | ||
| + | exit 1 | ||
| + | |||
| + | esac | ||
| + | </source> | ||
| + | |||
| + | |||
| + | |||
| + | :file1.txt: | ||
<pre style="display:block;font-family:monospace;white-space:pre;margin:1em 0;"> | <pre style="display:block;font-family:monospace;white-space:pre;margin:1em 0;"> | ||
| − | + | apples | |
| + | oranges | ||
| + | kiwis | ||
| + | carrots | ||
| + | </pre> | ||
| + | :file2.txt: | ||
| + | <pre style="display:block;font-family:monospace;white-space:pre;margin:1em 0;"> | ||
| + | apples | ||
| + | kiwis | ||
| + | carrots | ||
| + | grapefruits | ||
</pre> | </pre> | ||
| − | : | + | :diff -u result: |
| + | <pre style="display:block;font-family:monospace;white-space:pre;margin:1em 0;"> | ||
| + | --- file1.txt 2017-10-20 11:28:37.750414863 +0200 | ||
| + | +++ file2.txt 2017-10-20 11:28:54.638372491 +0200 | ||
| + | @@ -1,4 +1,4 @@ | ||
| + | apples | ||
| + | -oranges | ||
| + | kiwis | ||
| + | carrots | ||
| + | +grapefruits | ||
| + | </pre> | ||
| + | |||
| + | :sdiff -a result: | ||
| + | <pre style="display:block;font-family:monospace;white-space:pre;margin:1em 0;"> | ||
| + | apples apples | ||
| + | oranges < | ||
| + | kiwis kiwis | ||
| + | carrots carrots | ||
| + | > grapefruits | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | |||
| + | |||
| + | :DiffBasic code: | ||
[[File:DiffBasic.zip|DiffBasic.zip]] | [[File:DiffBasic.zip|DiffBasic.zip]] | ||
| + | |||
| + | == How to use specific Conda Environment == | ||
| + | To activate and use a specific environment is necessary to declare it in the code: | ||
| + | |||
| + | <source lang='bash'> | ||
| + | |||
| + | #!/bin/bash | ||
| + | echo "Ear_Detection" | ||
| + | echo "Parameter: " $1 | ||
| + | echo "Output: results.png" | ||
| + | |||
| + | source /home/gcube/conda2/bin/activate eartrack | ||
| + | python ear_detection.py $1 | ||
| + | source /home/gcube/conda2/bin/deactivate eartrack | ||
| + | |||
| + | |||
| + | </source> | ||
[[Category:Statistical Algorithms Importer]] | [[Category:Statistical Algorithms Importer]] | ||
Latest revision as of 11:44, 3 February 2021
F.A.Q. of Statistical Algorithms Importer (SAI), here are common mistakes we have found in Pre-Installed Project.
Windows User
Warning, this type of project will be launched in Linux environment. So be careful, all script files need to have a compatible Linux encoding. For example one difference is that Windows uses \r\n, whereas Linux uses \n for line termination. Another example is that Windows support "Unicode" UTF-16LE, and each character is 2 or 4 bytes, whereas Linux uses UTF-8, and each character is between 1 and 4 bytes. Also be aware of the Cut and Paste operation done from Windows machines that can enter unwanted \r\n characters. To solve this problem you can use the "convert to unix" functionality provided by notepad++ for example, or look for this feature in your editor.
How to use the input and output parameters
- For example we consider DiffBasic algorithm:
- DataMiner show the DiffBasic algorithm in this way:
- Bash script code in sample:
#!/bin/bash # # author: Giancarlo Panichi # echo "DiffBasic Project" echo "Author: Giancarlo Panchi" echo "Parameter operator: " $1 echo "Parameter options: " $2 echo "Parameter file1: " $3 echo "Parameter file2: " $4 echo "Output file: output.txt" OUT="output.txt" case "$1" in diff) diff $2 $3 $4 > $OUT ;; sdiff) sdiff $2 $3 $4 > $OUT ;; *) echo $"Usage: $1 {diff|sdiff}" exit 1 esac
- file1.txt:
apples oranges kiwis carrots
- file2.txt:
apples kiwis carrots grapefruits
- diff -u result:
--- file1.txt 2017-10-20 11:28:37.750414863 +0200 +++ file2.txt 2017-10-20 11:28:54.638372491 +0200 @@ -1,4 +1,4 @@ apples -oranges kiwis carrots +grapefruits
- sdiff -a result:
apples apples
oranges <
kiwis kiwis
carrots carrots
> grapefruits
- DiffBasic code:
How to use specific Conda Environment
To activate and use a specific environment is necessary to declare it in the code:
#!/bin/bash echo "Ear_Detection" echo "Parameter: " $1 echo "Output: results.png" source /home/gcube/conda2/bin/activate eartrack python ear_detection.py $1 source /home/gcube/conda2/bin/deactivate eartrack