Difference between revisions of "Statistical Algorithms Importer: FAQ"
(→Project Name) |
|||
| (48 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
|} | |} | ||
| − | F.A.Q. of Statistical Algorithms Importer (SAI), here are common mistakes we have found. | + | F.A.Q. of [[Statistical_Algorithms_Importer|Statistical Algorithms Importer (SAI)]], here are common mistakes we have found. |
| − | == | + | == Project Type FAQ == |
| − | + | * [[Statistical Algorithms Importer: R Project FAQ|R Project FAQ]] | |
| + | * [[Statistical Algorithms Importer: R-blackbox Project FAQ|R-blackbox Project FAQ]] | ||
| + | * [[Statistical Algorithms Importer: Java Project FAQ|Java Project FAQ]] | ||
| + | * [[Statistical Algorithms Importer: Linux-compiled Project FAQ|Linux-compiled Project FAQ]] | ||
| + | * [[Statistical Algorithms Importer: Python Project FAQ|Python Project FAQ]] | ||
| + | * [[Statistical Algorithms Importer: Pre-Installed Project FAQ|Pre-Installed Project FAQ]] | ||
| − | * | + | == Installed Software == |
| + | A list of pre-installed software on the infrastructure machines is available at this page: | ||
| + | * [[Pre Installed Packages|Pre Installed Packages]] | ||
| − | + | In general it is better to specify the packages with the relative versions as they shown in the previous link. | |
| − | + | However, if you do not specify the packages the system tries to integrate and run the code using the packages already present on the DataMiner, this is done to facilitate the integration of the developers. | |
| − | + | Obviously, in this case if the process uses non-installed packages it will fail during the execution and it will be the developer's responsibility to request the installation of the missing packages. | |
| − | + | The Interpreter version also serves to better identify the type of code being executed and to support the entire debugging phase in the event of problems. | |
| + | So in general, algorithm support will be better if the information generated is greater, but in any case the system tries to integrate and execute the code. | ||
| − | + | == Project Folder == | |
| − | + | It is important that each algorithm has its own project folder. The project folder keeps the code created by the developer, so it is important that each algorithm has its own project folder, different for each algorithm. Once an algorithm is published, the Project Folder will contain the executable that will be requested by the DataMiner for execution, so it is important to avoid deleting published projects. Deleting a project means to establish to make it unavailable for use in the infrastructure. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | == Project Name == | |
| + | The project name cannot contain special characters, only letters and numbers are allowed, moreover any spaces can be replaced by the underscore character. Each project must have its own name different from that used in other projects. | ||
| − | + | Project names already used: | |
| − | : | + | * [[DataMiner_Project_Names_Already_Used|DataMiner Project Names Already Used]] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | == | + | == Project Configuration == |
| − | + | The SAI uses two project configuration files: | |
| + | * stat_algo.project | ||
| + | * Main.R | ||
| + | It is advisable that these files are never deleted or modified directly. | ||
| − | + | == Project ID == | |
| + | Starting from the project name a unique identifier is associated to each project when it is published. | ||
| + | The identifier allows the project to be recognized within the infrastructure. | ||
| + | This is why it is important to give different names to each project and not to reuse the same name in different projects. | ||
| − | + | Where can find the Project Id? Just check the link associated with the algorithm name in DataMiner. | |
| − | + | [[Image:StatisticalAlgorithmsImporter_ProjectID.png|thumb|center|750px|Project ID, SAI]] | |
| − | + | ||
| − | + | ||
| − | + | == Parameters == | |
| − | + | It is important that an algorithm always has at least one input and one output parameter. | |
| − | + | All parameters are mandatory this is a design choice to support the repeatability and the reproducibility of the experiments, and also the reuse of algorithms. | |
| − | + | In the case you want to include one optional file, it would be better to create two distinct algorithms, one that expects the file parameter and the other that does not. | |
| − | + | Of course you can use default value in the case of Strings, Integers, .... etcetera. | |
| − | + | == I don't see my algorithm in DataMiner == | |
| − | + | DataMiner portlets store algorithms in the user session, so if an algorithm is deployed but is not visible you must try to refresh the list of algorithms with the refresh button in the DataMiner. | |
| − | + | Remember, after the deploy a few minutes are needed to upgrade the system. | |
| − | + | ||
| − | + | ||
| − | + | == Publish an algorithm the first time == | |
| + | The first time an algorithm is created, it must be published using the Publish button in the current VRE. After the first publication, both Repackage and Publish can be used. | ||
| + | In the case that the Input and Output parameters are changed then it is necessary to reuse the Publish. | ||
| − | == | + | == Publish in another VRE == |
| − | + | Sometimes we want to publish an algorithm in another VRE, different from the one in which we have already published the algorithm. | |
| + | If the SAI is present in the new VRE, just open the algorithm in the new VRE and publish it, otherwise you can open a ticket and you can report the VRE and the name of the algorithm that you want to publish. | ||
| − | + | == Delete an algorithm == | |
| + | To delete an algorithm published through the SAI it is necessary to open a ticket. The name of the algorithm and the list of VREs in which it was published must be written in the ticket. | ||
| − | + | == Advanced Input == | |
| − | + | It is possible to indicate spatial inputs or time/date inputs. The details for the definition of these dare are reported in the [[Advanced Input| Advanced Input ]] | |
| − | + | ||
| − | == | + | == Update the status of a computation == |
| − | + | It is possible to update the inner status of a computation by writing a status.txt file locally to the process [[Statistical Algorithms Importer: StatusUpdate| Updating the status of a computation]] | |
| − | + | ||
| − | + | == Docker Support == | |
| − | + | :SAI and DataMiner support the execution of Docker images on D4Science, for more information see the wiki available at this page: | |
| − | + | * [[Statistical Algorithms Importer: Docker Support|Statistical Algorithms Importer: Docker Support]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | = | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
[[Category:Statistical Algorithms Importer]] | [[Category:Statistical Algorithms Importer]] | ||
Latest revision as of 18:26, 24 March 2021
F.A.Q. of Statistical Algorithms Importer (SAI), here are common mistakes we have found.
Project Type FAQ
- R Project FAQ
- R-blackbox Project FAQ
- Java Project FAQ
- Linux-compiled Project FAQ
- Python Project FAQ
- Pre-Installed Project FAQ
Installed Software
A list of pre-installed software on the infrastructure machines is available at this page:
In general it is better to specify the packages with the relative versions as they shown in the previous link. However, if you do not specify the packages the system tries to integrate and run the code using the packages already present on the DataMiner, this is done to facilitate the integration of the developers. Obviously, in this case if the process uses non-installed packages it will fail during the execution and it will be the developer's responsibility to request the installation of the missing packages. The Interpreter version also serves to better identify the type of code being executed and to support the entire debugging phase in the event of problems. So in general, algorithm support will be better if the information generated is greater, but in any case the system tries to integrate and execute the code.
Project Folder
It is important that each algorithm has its own project folder. The project folder keeps the code created by the developer, so it is important that each algorithm has its own project folder, different for each algorithm. Once an algorithm is published, the Project Folder will contain the executable that will be requested by the DataMiner for execution, so it is important to avoid deleting published projects. Deleting a project means to establish to make it unavailable for use in the infrastructure.
Project Name
The project name cannot contain special characters, only letters and numbers are allowed, moreover any spaces can be replaced by the underscore character. Each project must have its own name different from that used in other projects.
Project names already used:
Project Configuration
The SAI uses two project configuration files:
- stat_algo.project
- Main.R
It is advisable that these files are never deleted or modified directly.
Project ID
Starting from the project name a unique identifier is associated to each project when it is published. The identifier allows the project to be recognized within the infrastructure. This is why it is important to give different names to each project and not to reuse the same name in different projects.
Where can find the Project Id? Just check the link associated with the algorithm name in DataMiner.
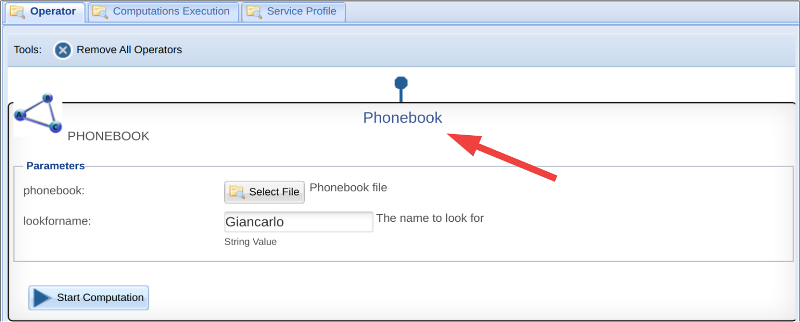
Parameters
It is important that an algorithm always has at least one input and one output parameter. All parameters are mandatory this is a design choice to support the repeatability and the reproducibility of the experiments, and also the reuse of algorithms. In the case you want to include one optional file, it would be better to create two distinct algorithms, one that expects the file parameter and the other that does not. Of course you can use default value in the case of Strings, Integers, .... etcetera.
I don't see my algorithm in DataMiner
DataMiner portlets store algorithms in the user session, so if an algorithm is deployed but is not visible you must try to refresh the list of algorithms with the refresh button in the DataMiner. Remember, after the deploy a few minutes are needed to upgrade the system.
Publish an algorithm the first time
The first time an algorithm is created, it must be published using the Publish button in the current VRE. After the first publication, both Repackage and Publish can be used. In the case that the Input and Output parameters are changed then it is necessary to reuse the Publish.
Publish in another VRE
Sometimes we want to publish an algorithm in another VRE, different from the one in which we have already published the algorithm. If the SAI is present in the new VRE, just open the algorithm in the new VRE and publish it, otherwise you can open a ticket and you can report the VRE and the name of the algorithm that you want to publish.
Delete an algorithm
To delete an algorithm published through the SAI it is necessary to open a ticket. The name of the algorithm and the list of VREs in which it was published must be written in the ticket.
Advanced Input
It is possible to indicate spatial inputs or time/date inputs. The details for the definition of these dare are reported in the Advanced Input
Update the status of a computation
It is possible to update the inner status of a computation by writing a status.txt file locally to the process Updating the status of a computation
Docker Support
- SAI and DataMiner support the execution of Docker images on D4Science, for more information see the wiki available at this page: