Difference between revisions of "Biodiversity Access"
m (→Architecture) |
(→Small deployment) |
||
| (48 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <!-- CATEGORIES --> | ||
| + | [[Category: gCube Features]] | ||
| + | <!-- CATEGORIES --> | ||
| + | {| align="right" | ||
| + | ||__TOC__ | ||
| + | |} | ||
| + | Part of the [[Data Access and Storage Facilities]], a cluster of components within the system focus on uniform access to sources of biodiversity data of arbitrary location and size. | ||
| − | + | This document outlines their design rationale, key features, and high-level architecture, as well as the options for their deployment. | |
| − | This document outlines their design rationale, key features, and high-level architecture as well as the options for their deployment. | + | |
== Overview == | == Overview == | ||
The goal of this subsystem is to offer an uniform access over different biodiversity repositories through a simple API. | The goal of this subsystem is to offer an uniform access over different biodiversity repositories through a simple API. | ||
| − | The | + | |
| + | The service has dynamically extensible architecture, i.e. rely on independently developed plugins to adapt their APIs to a variety of back-ends within or outside the system. | ||
| + | |||
When connected to remote data sources, the services may be widely replicated and their replicas know how to leverage the Enabling Services to scale horizontally to the capacity of the remote back-ends. | When connected to remote data sources, the services may be widely replicated and their replicas know how to leverage the Enabling Services to scale horizontally to the capacity of the remote back-ends. | ||
| + | === Key features === | ||
| + | |||
| + | * [[Data_Access_and_Storage_APIs#SPD_Client_API | access API tailored to biodiversity data sources]]; | ||
| + | * unifying [[Species_Products_Discovery_Objects | object model for Species Products]]; | ||
| + | * dynamically pluggable architecture of transformations (eg unified classification, synonyms presentation) from external sources of biodiversity data; | ||
| + | * plugins for key biodiversity data sources, including OBIS, GBIF and Catalogue of Life; | ||
| + | * dynamic clustering of discovered items; | ||
| + | * integration with workspace facilities; | ||
| + | * export of discovered items in multiple formats, including DarwinCore and DarwinCore Archive; | ||
| + | * [[SPQL:_SPecies_Query_Language | flexible query language]]; | ||
== Design == | == Design == | ||
| Line 15: | Line 33: | ||
=== Philosophy === | === Philosophy === | ||
| − | + | Uniform access to sources of biodiversity data that expose different capabilities and different data models is a key requirement to support biodiversity studies. | |
| − | This subsystem offer the possibility to retrieve, | + | |
| − | + | This subsystem offer the possibility to retrieve, manage and elaborate biodiversity data under a single model, with a domain-specific API, and regardless of its source of origin. | |
=== Architecture === | === Architecture === | ||
| − | + | The subsystem comprises the following components: | |
| + | |||
| + | * '''species-products-discovery-service''': a stateless Web Service that exposes read operations and implements it by delegation to dynamically deployable plugins for target repository sources within and outside the system; | ||
| + | |||
| + | * '''species-products-discovery-library''': a client library that implements a high-level facade to the remote APIs of the Species manager service; | ||
| + | |||
| + | * '''obis-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.iobis.org OBIS] data source; | ||
| + | |||
| + | * '''gbif-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.gbif.org GBIF] data source; | ||
| + | |||
| + | * '''catalogueoflife-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.catalogueoflife.org Catalogue of Life] data source; | ||
| + | |||
| + | * '''flora-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://floradobrasil.jbrj.gov.br Brazilian Flora] data source. | ||
| + | |||
| + | * '''worms-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.marinespecies.org/ Worms] data source. | ||
| + | |||
| + | * '''wordss-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.marinespecies.org/deepsea/ World Register of Deep Sea Species] data source. | ||
| + | |||
| + | * '''species-link-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://splink.cria.org.br/ speciesLink] data source, by using the international standard TAPIR protocol. | ||
| + | |||
| + | * '''itis-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.itis.gov/ ITIS] data source. | ||
| + | |||
| + | * '''ncbi-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.ncbi.nlm.nih.gov/ NCBI] data source. | ||
| + | |||
| + | * '''irmng-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.cmar.csiro.au/datacentre/irmng/ IRMNG] data source, through Darwin Core Archive format. | ||
| + | |||
| + | * '''asfis-spd-plugin''': a plugin of the Species Products Discovery service that interacts with [http://www.fao.org/fishery/collection/asfis/en ASFIS] data source. | ||
| + | |||
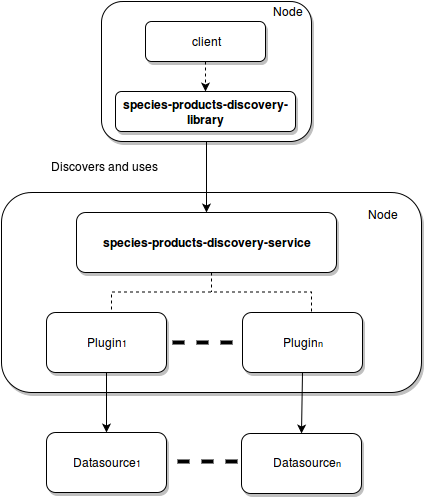
| + | A diagram of the relationships between these components is reported in the following figure: | ||
| + | |||
| + | |||
| + | [[Image:species-manager-arch_new.png|frame|center|Biodiversity Access Subsystem Architecture]] | ||
| + | |||
| + | == Deployment == | ||
| + | |||
| + | All the components of the subsystem must be deployed together in a single node. | ||
| + | This subsystem can be replicated in multiple hosts; this does not guarantee a performance improvement because the scalability of this system depends on the capacity of external repositories contacted by the plugins. | ||
| + | There are no temporal constraints on the co-deployment of services and plugins. Every plugin must be deployed on every instance of the service. | ||
| + | This subsystem is lightweight, it does not need of excessive memory or disk space. | ||
| + | |||
| + | |||
| − | + | === Small deployment === | |
| − | + | [[Image:species-manager-small-depl_new.png|frame|center|Biodiversity Access Deployment]] | |
| − | + | == Use Cases == | |
| − | + | === Well suited Use Cases === | |
| − | + | The subsystem is particularly suited to support abstraction over biodiversity data. Every biodiversity repository can be easily integrated in this subsystem developing a plugin. | |
| − | + | The development of any plugin of the Species Manager services immediately extends the ability of the systems to discovery new biodiversity data. | |
Latest revision as of 16:28, 6 June 2016
Part of the Data Access and Storage Facilities, a cluster of components within the system focus on uniform access to sources of biodiversity data of arbitrary location and size.
This document outlines their design rationale, key features, and high-level architecture, as well as the options for their deployment.
Overview
The goal of this subsystem is to offer an uniform access over different biodiversity repositories through a simple API.
The service has dynamically extensible architecture, i.e. rely on independently developed plugins to adapt their APIs to a variety of back-ends within or outside the system.
When connected to remote data sources, the services may be widely replicated and their replicas know how to leverage the Enabling Services to scale horizontally to the capacity of the remote back-ends.
Key features
- access API tailored to biodiversity data sources;
- unifying object model for Species Products;
- dynamically pluggable architecture of transformations (eg unified classification, synonyms presentation) from external sources of biodiversity data;
- plugins for key biodiversity data sources, including OBIS, GBIF and Catalogue of Life;
- dynamic clustering of discovered items;
- integration with workspace facilities;
- export of discovered items in multiple formats, including DarwinCore and DarwinCore Archive;
- flexible query language;
Design
Philosophy
Uniform access to sources of biodiversity data that expose different capabilities and different data models is a key requirement to support biodiversity studies.
This subsystem offer the possibility to retrieve, manage and elaborate biodiversity data under a single model, with a domain-specific API, and regardless of its source of origin.
Architecture
The subsystem comprises the following components:
- species-products-discovery-service: a stateless Web Service that exposes read operations and implements it by delegation to dynamically deployable plugins for target repository sources within and outside the system;
- species-products-discovery-library: a client library that implements a high-level facade to the remote APIs of the Species manager service;
- obis-spd-plugin: a plugin of the Species Products Discovery service that interacts with OBIS data source;
- gbif-spd-plugin: a plugin of the Species Products Discovery service that interacts with GBIF data source;
- catalogueoflife-spd-plugin: a plugin of the Species Products Discovery service that interacts with Catalogue of Life data source;
- flora-spd-plugin: a plugin of the Species Products Discovery service that interacts with Brazilian Flora data source.
- worms-spd-plugin: a plugin of the Species Products Discovery service that interacts with Worms data source.
- wordss-spd-plugin: a plugin of the Species Products Discovery service that interacts with World Register of Deep Sea Species data source.
- species-link-spd-plugin: a plugin of the Species Products Discovery service that interacts with speciesLink data source, by using the international standard TAPIR protocol.
- itis-spd-plugin: a plugin of the Species Products Discovery service that interacts with ITIS data source.
- ncbi-spd-plugin: a plugin of the Species Products Discovery service that interacts with NCBI data source.
- irmng-spd-plugin: a plugin of the Species Products Discovery service that interacts with IRMNG data source, through Darwin Core Archive format.
- asfis-spd-plugin: a plugin of the Species Products Discovery service that interacts with ASFIS data source.
A diagram of the relationships between these components is reported in the following figure:
Deployment
All the components of the subsystem must be deployed together in a single node. This subsystem can be replicated in multiple hosts; this does not guarantee a performance improvement because the scalability of this system depends on the capacity of external repositories contacted by the plugins. There are no temporal constraints on the co-deployment of services and plugins. Every plugin must be deployed on every instance of the service. This subsystem is lightweight, it does not need of excessive memory or disk space.
Small deployment
Use Cases
Well suited Use Cases
The subsystem is particularly suited to support abstraction over biodiversity data. Every biodiversity repository can be easily integrated in this subsystem developing a plugin.
The development of any plugin of the Species Manager services immediately extends the ability of the systems to discovery new biodiversity data.